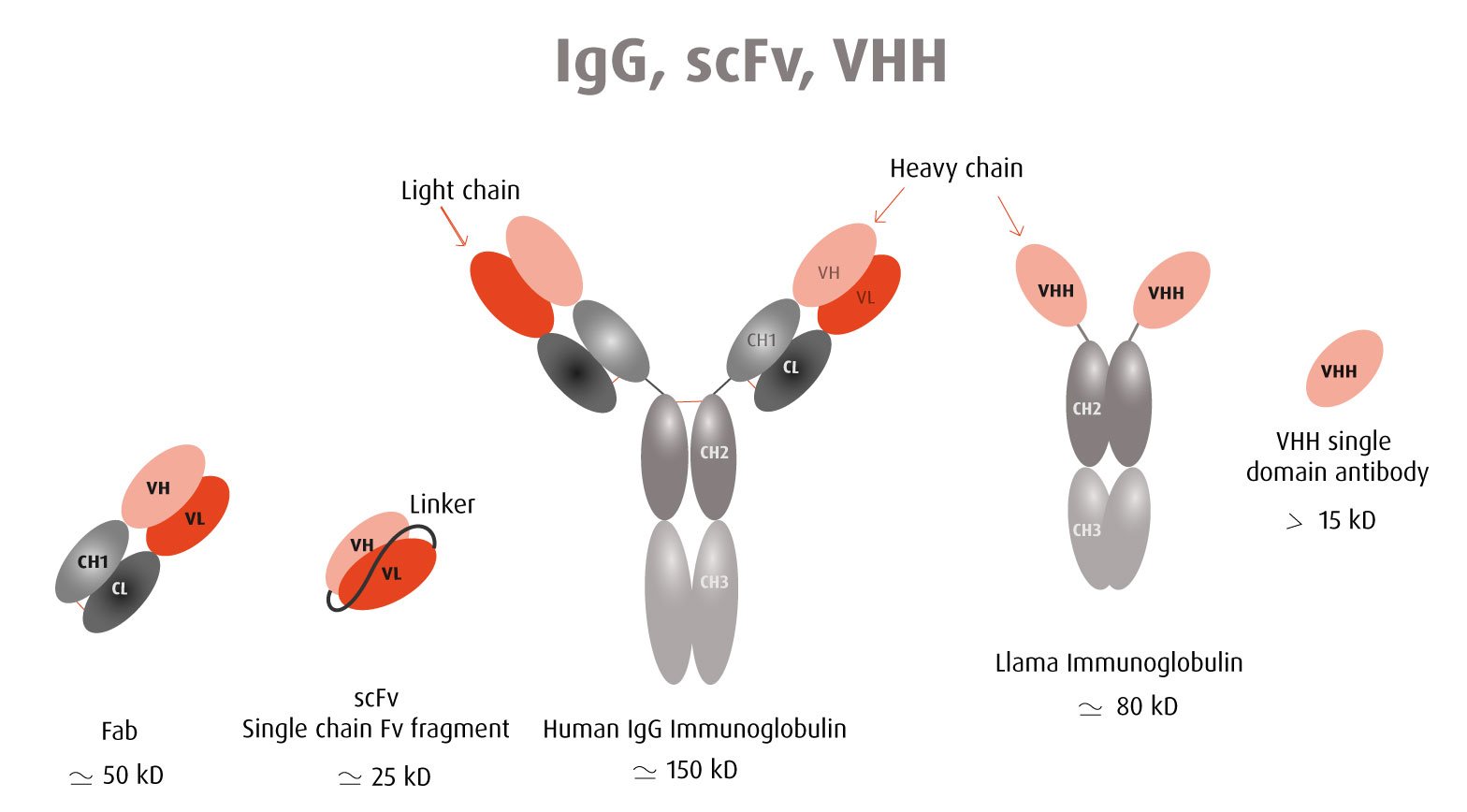
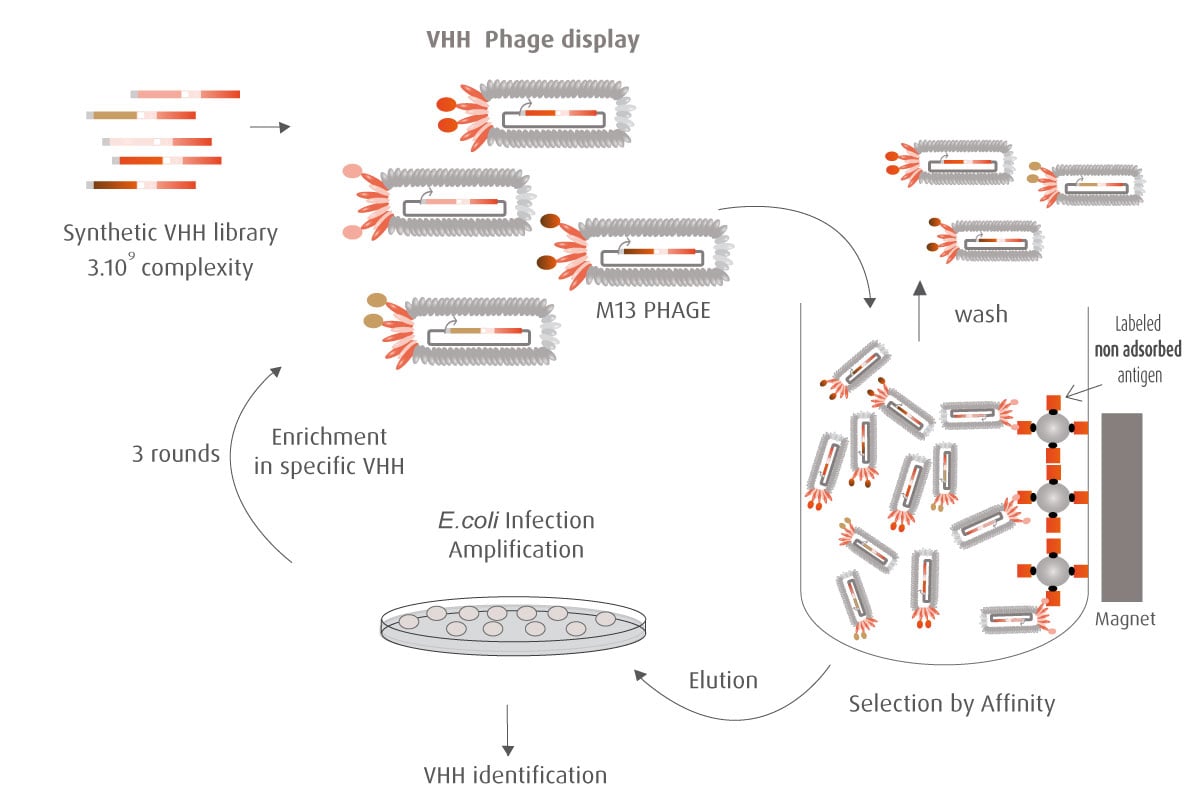
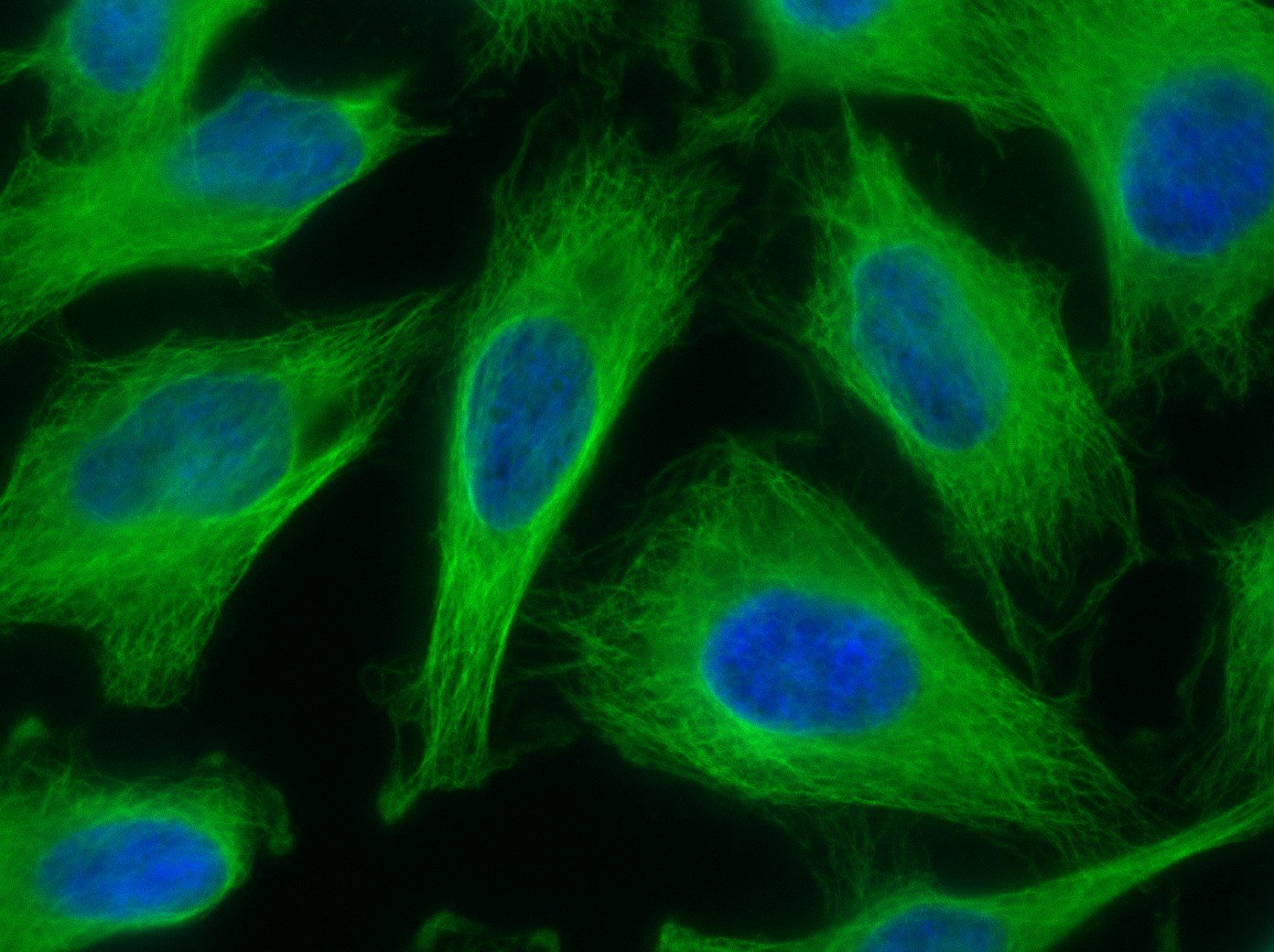
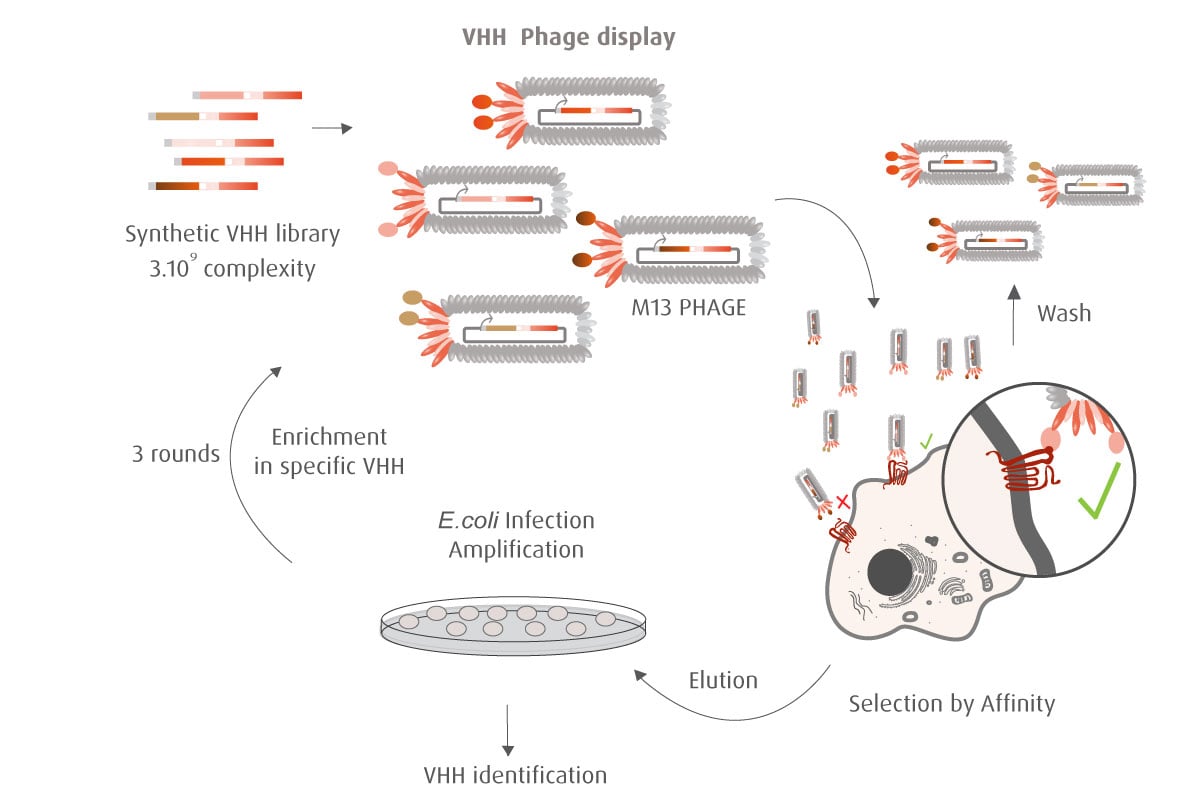
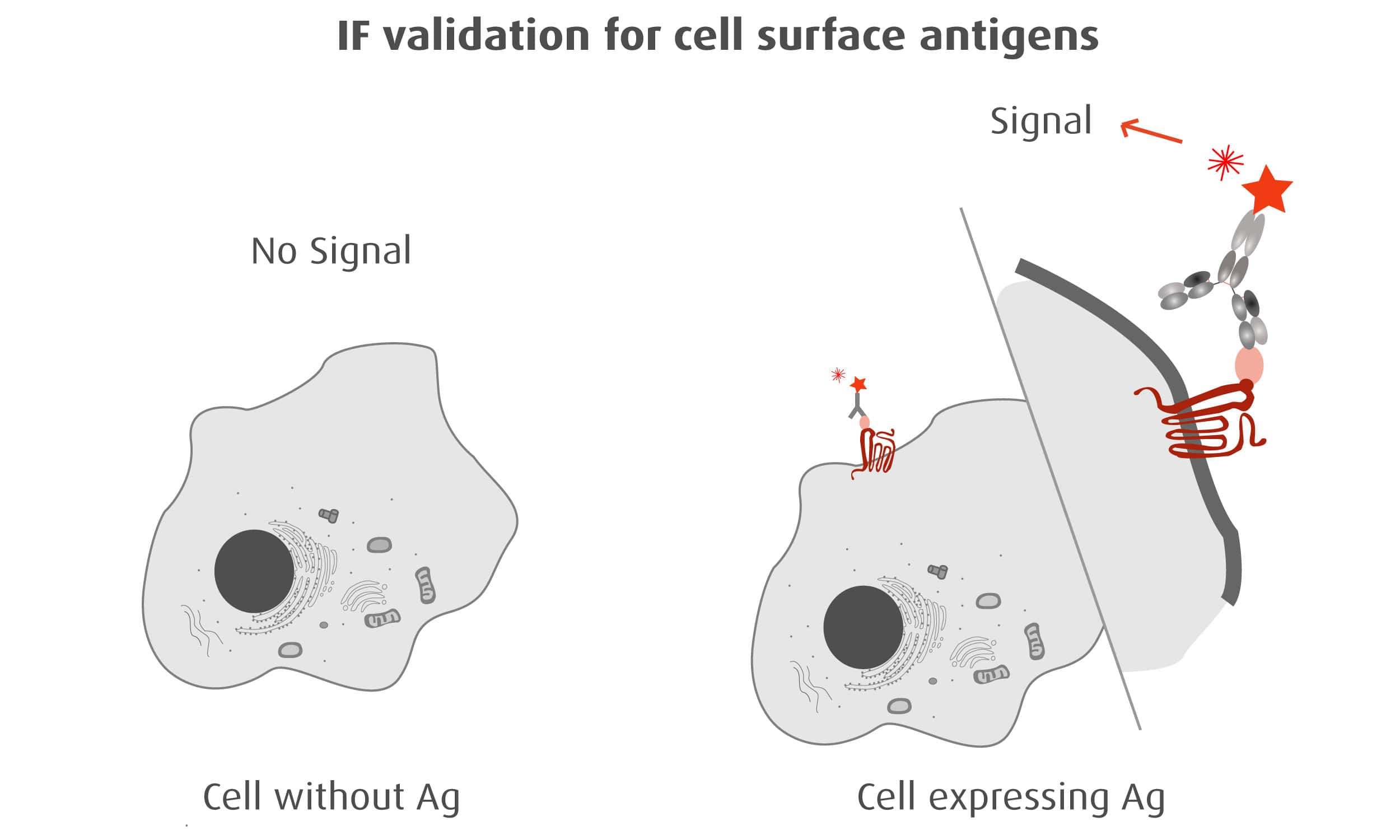
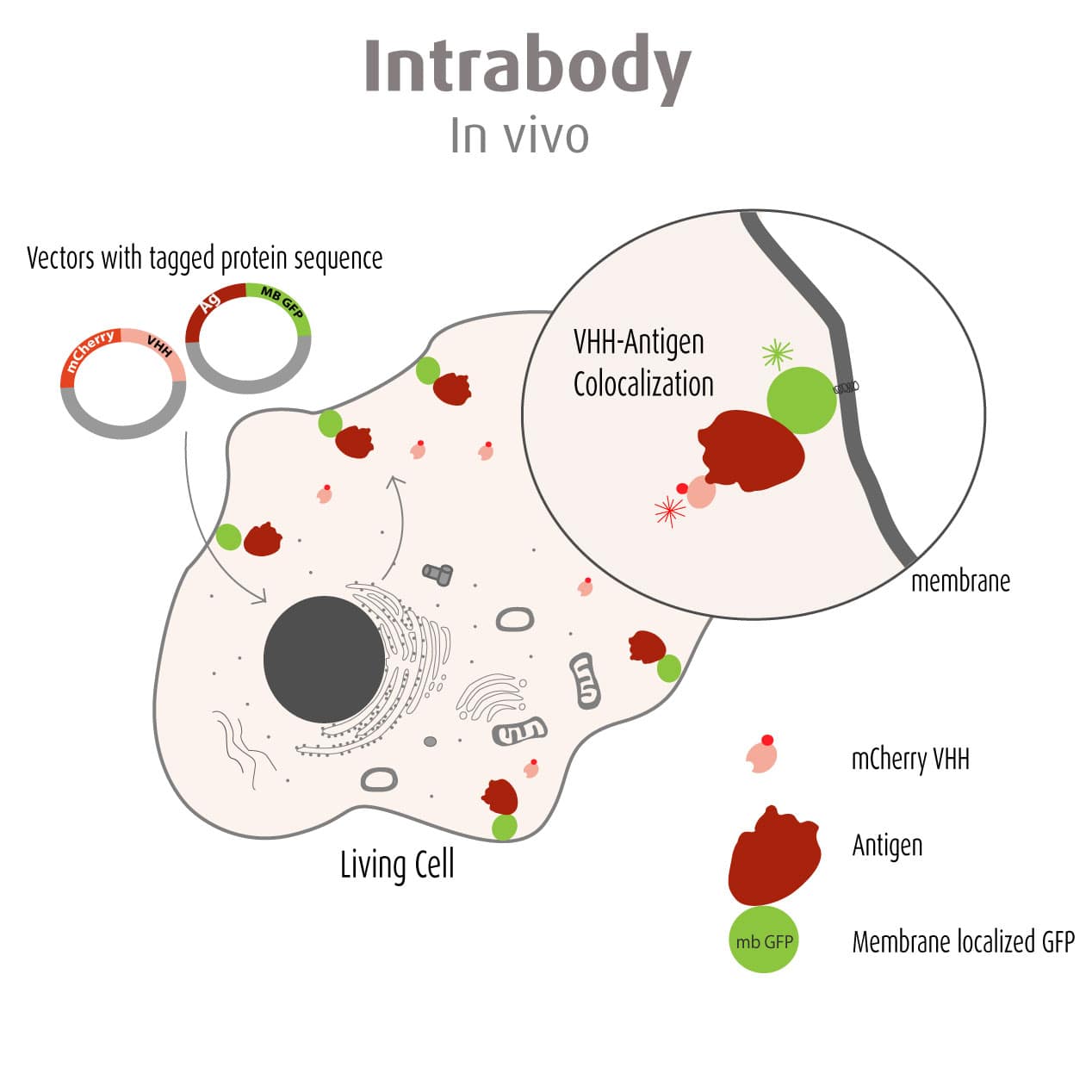
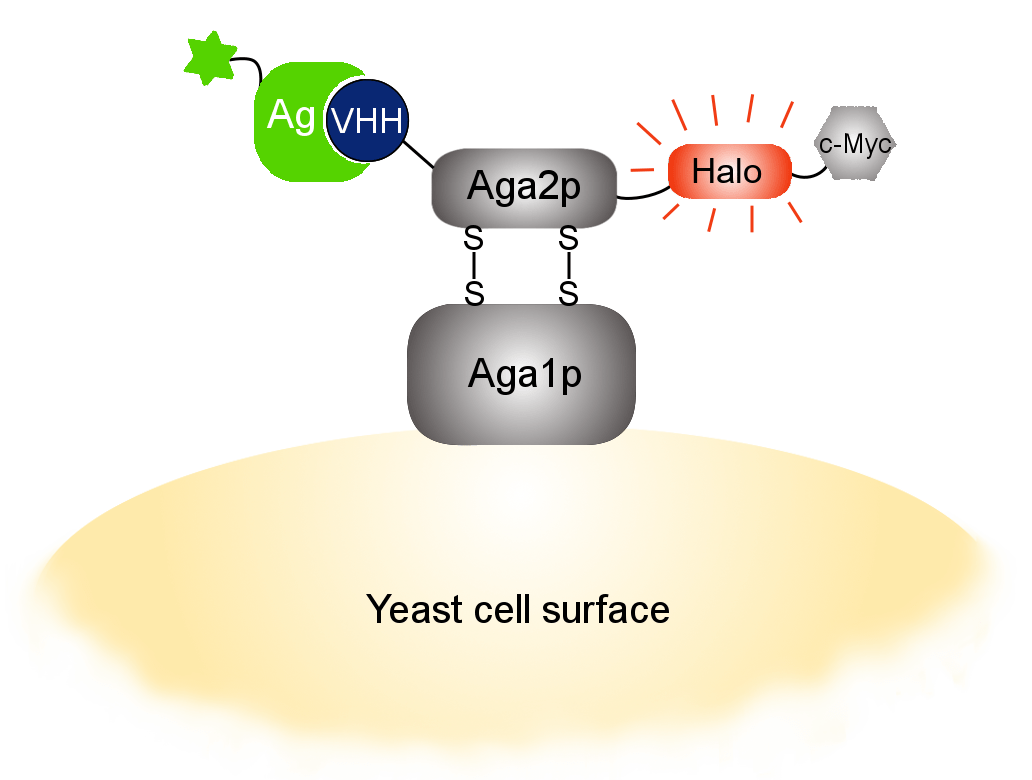
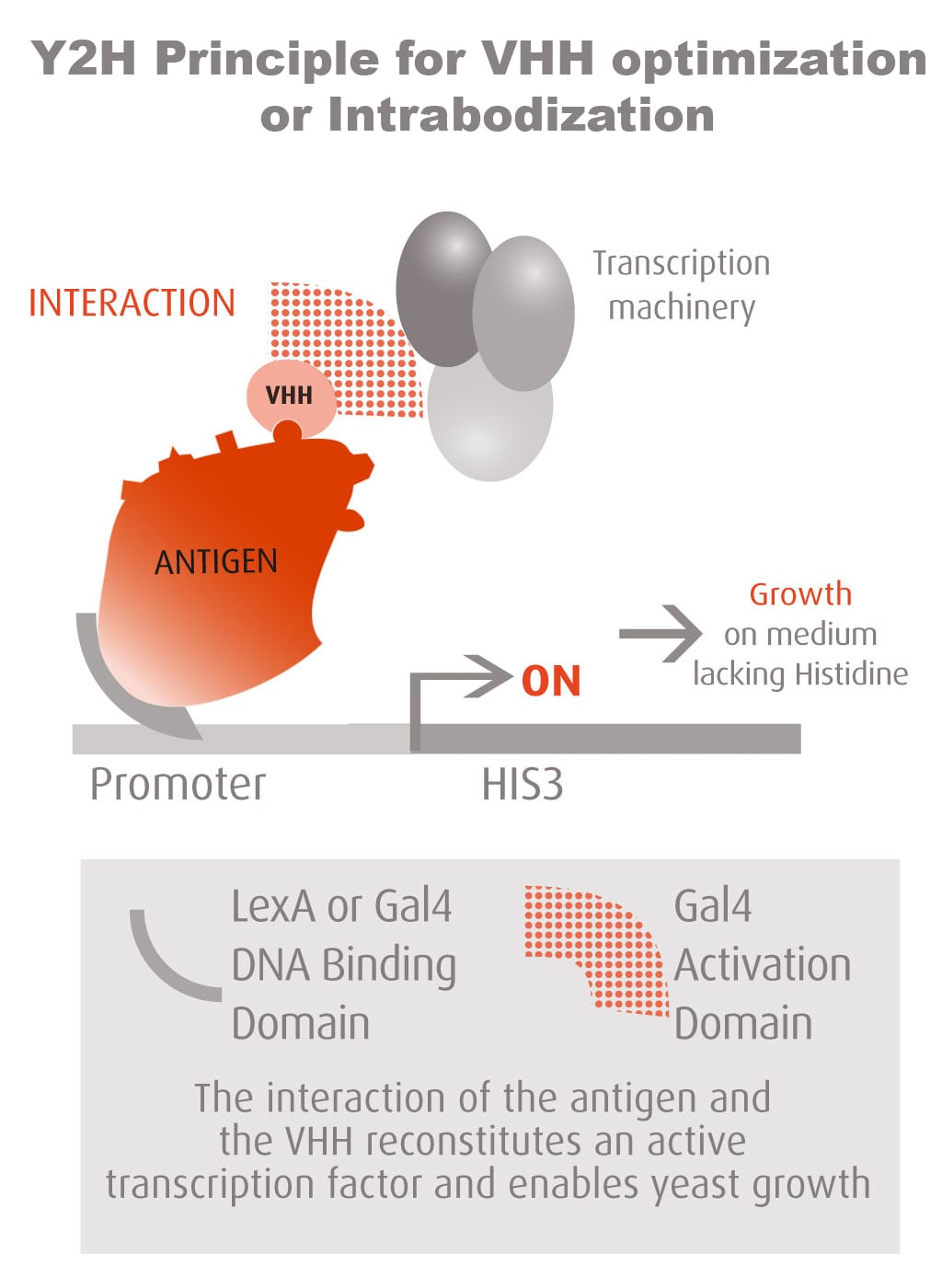
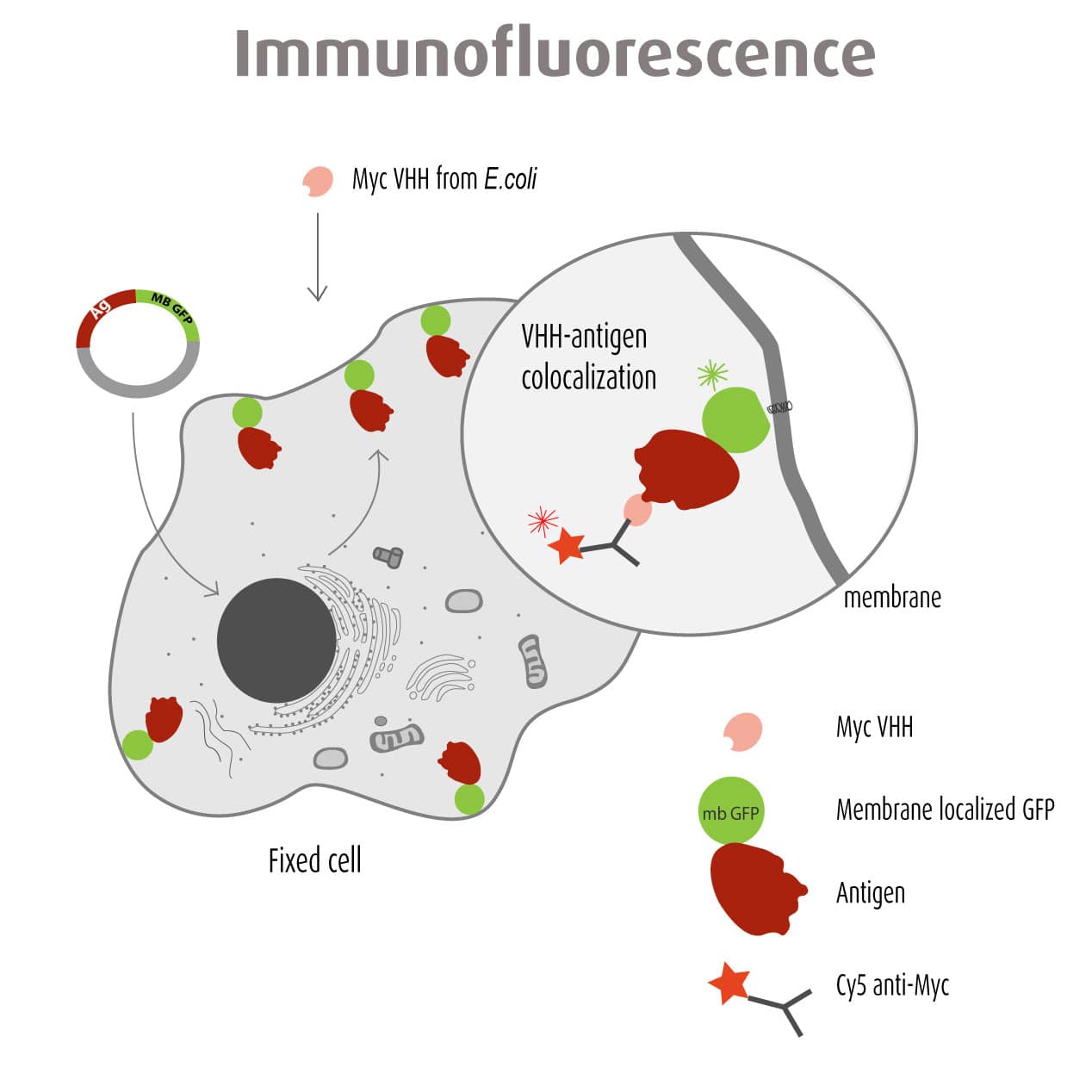
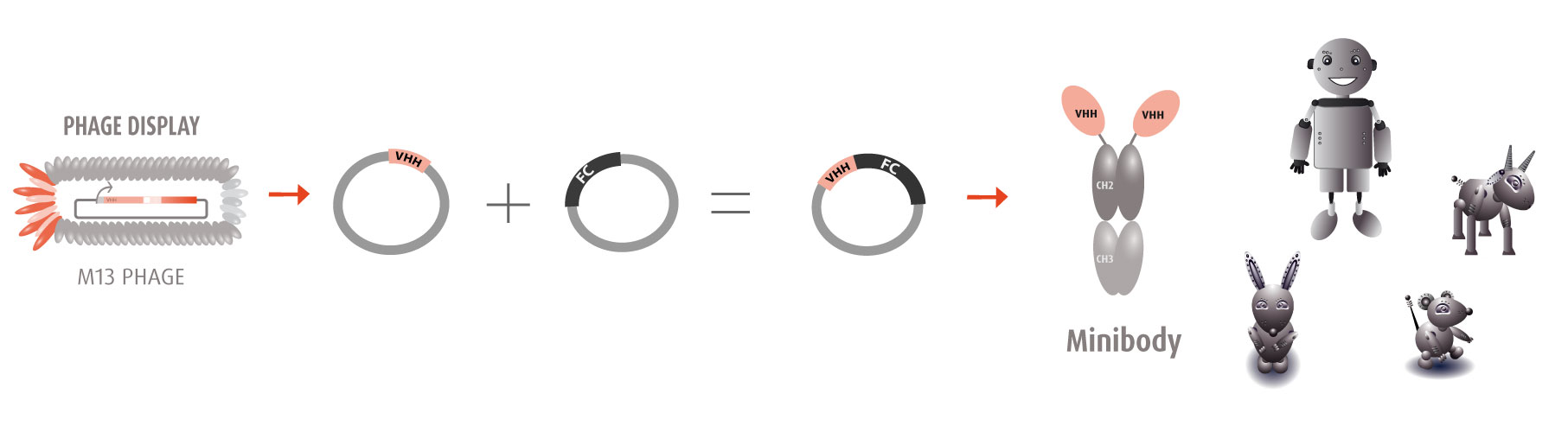Our optimized Phage Display selection technology delivers high quality and fully functional humanized recombinant nanobodies recognizing native antigens. Importantly, the structural integrity of the antigen is maintained throughout the entire VHH selection and validation process. Therefore, our nanobodies are more likely to recognize endogenous antigens for in vitro and in vivo applications, such as immunofluorescence, live cell imaging, protein degradation or blocking protein activity.
Key benefits
- Conformational antibodies
- Deliverables of the validated VHH binders include DNA sequences and plasmids
- Customized selection conditions (buffer, temperature, etc)
- Animal-free technology